Initial Structure to Relaxed Structure (IS2RS)
Contents
Initial Structure to Relaxed Structure (IS2RS)#
We approach the IS2RS task by using a pre-trained S2EF model to iteratively run a structure optimization to arrive at a relaxed structure. While the majority of approaches for this task do this iteratively, we note it’s possible to train a model to directly predict relaxed structures.
Steps for making IS2RS predictions#
Define or load a configuration (config), which includes the following
task with relaxation dataset information
model
optimizer
dataset
trainer
Create a ForcesTrainer object
Train a S2EF model or load an existing S2EF checkpoint
Run relaxations
Note For this task we’ll be using a publicly released pre-trained checkpoint of our best model to perform relaxations.
Imports#
from ocpmodels import models
from ocpmodels.common import logger
from ocpmodels.common.utils import setup_logging
from ocpmodels.datasets import TrajectoryLmdbDataset
from ocpmodels.trainers import ForcesTrainer
setup_logging()
import numpy as np
Dataset#
The IS2RS task requires an additional relaxation dataset to be defined - relax_dataset. This dataset is read in similar to the IS2RE dataset - requiring an LMDB file. The same datasets are used for the IS2RE and IS2RS tasks.
%%bash
mkdir data
cd data
wget -q -nc http://dl.fbaipublicfiles.com/opencatalystproject/data/tutorial_data.tar.gz -O tutorial_data.tar.gz
tar -xzvf tutorial_data.tar.gz
mkdir: cannot create directory ‘data’: File exists
./
./is2re/
./is2re/train_100/
./is2re/train_100/data.lmdb
./is2re/train_100/data.lmdb-lock
./is2re/val_20/
./is2re/val_20/data.lmdb
./is2re/val_20/data.lmdb-lock
./s2ef/
./s2ef/train_100/
./s2ef/train_100/data.lmdb
./s2ef/train_100/data.lmdb-lock
./s2ef/val_20/
./s2ef/val_20/data.lmdb
./s2ef/val_20/data.lmdb-lock
train_src = "data/s2ef/train_100"
val_src = "data/s2ef/val_20"
relax_dataset = "data/is2re/val_20/data.lmdb"
Download pretrained checkpoint#
!wget -q https://dl.fbaipublicfiles.com/opencatalystproject/models/2021_08/s2ef/gemnet_t_direct_h512_all.pt
checkpoint_path = "/content/ocp/gemnet_t_direct_h512_all.pt"
Define the Config#
Running an iterative S2EF model for the IS2RS task can be run from any S2EF config given the following additions to the task portion of the config:
relax_dataset - IS2RE LMDB dataset
write_pos - Whether to save out relaxed positions
relaxation_steps - Number of optimization steps to run
relax_opt - Dictionary of optimizer settings. Currently only LBFGS supported
maxstep - Maximum distance an optimization is allowed to make
memory - Memory history to use for LBFGS
damping - Calculated step is multiplied by this factor before updating positions
alpha - Initial guess for the Hessian
traj_dir - If specified, directory to save out the full ML relaxation as an ASE trajectory. Useful for debugging or visualizing results.
num_relaxation_batches - If specified, relaxations will only be run for a subset of the relaxation dataset. Useful for debugging or wanting to visualize a few systems.
A sample relaxation config can be found here.
# Task
task = {
"dataset": "trajectory_lmdb", # dataset used for the S2EF task
"description": "Regressing to energies and forces for DFT trajectories from OCP",
"type": "regression",
"metric": "mae",
"labels": ["potential energy"],
"grad_input": "atomic forces",
"train_on_free_atoms": True,
"eval_on_free_atoms": True,
"relax_dataset": {"src": relax_dataset},
"write_pos": True,
"relaxation_steps": 200,
"num_relaxation_batches": 1,
"relax_opt": {
"maxstep": 0.04,
"memory": 50,
"damping": 1.0,
"alpha": 70.0,
"traj_dir": "ml-relaxations/is2rs-test",
},
}
# Model
model = {
"name": "gemnet_t",
"num_spherical": 7,
"num_radial": 128,
"num_blocks": 3,
"emb_size_atom": 512,
"emb_size_edge": 512,
"emb_size_trip": 64,
"emb_size_rbf": 16,
"emb_size_cbf": 16,
"emb_size_bil_trip": 64,
"num_before_skip": 1,
"num_after_skip": 2,
"num_concat": 1,
"num_atom": 3,
"cutoff": 6.0,
"max_neighbors": 50,
"rbf": {"name": "gaussian"},
"envelope": {
"name": "polynomial",
"exponent": 5,
},
"cbf": {"name": "spherical_harmonics"},
"extensive": True,
"otf_graph": False,
"output_init": "HeOrthogonal",
"activation": "silu",
"scale_file": "configs/s2ef/all/gemnet/scaling_factors/gemnet-dT.json",
"regress_forces": True,
"direct_forces": True,
}
# Optimizer
optimizer = {
"batch_size": 1, # originally 32
"eval_batch_size": 1, # originally 32
"num_workers": 2,
"lr_initial": 5.0e-4,
"optimizer": "AdamW",
"optimizer_params": {"amsgrad": True},
"scheduler": "ReduceLROnPlateau",
"mode": "min",
"factor": 0.8,
"ema_decay": 0.999,
"clip_grad_norm": 10,
"patience": 3,
"max_epochs": 1, # used for demonstration purposes
"force_coefficient": 100,
}
# Dataset
dataset = [
{"src": train_src, "normalize_labels": False}, # train set
{"src": val_src}, # val set (optional)
]
Create the trainer#
trainer = ForcesTrainer(
task=task,
model=model,
dataset=dataset,
optimizer=optimizer,
identifier="is2rs-example",
run_dir="./", # directory to save results if is_debug=False. Prediction files are saved here so be careful not to override!
is_debug=False, # if True, do not save checkpoint, logs, or results
print_every=5,
seed=0, # random seed to use
logger="tensorboard", # logger of choice (tensorboard and wandb supported)
local_rank=0,
amp=True, # use PyTorch Automatic Mixed Precision (faster training and less memory usage)
)
amp: true
cmd:
checkpoint_dir: ./checkpoints/2022-10-31-18-01-36-is2rs-example
commit: cba9fb6
identifier: is2rs-example
logs_dir: ./logs/tensorboard/2022-10-31-18-01-36-is2rs-example
print_every: 5
results_dir: ./results/2022-10-31-18-01-36-is2rs-example
seed: 0
timestamp_id: 2022-10-31-18-01-36-is2rs-example
dataset:
normalize_labels: false
src: data/s2ef/train_100
gpus: 0
logger: tensorboard
model: gemnet_t
model_attributes:
activation: silu
cbf:
name: spherical_harmonics
cutoff: 6.0
direct_forces: true
emb_size_atom: 512
emb_size_bil_trip: 64
emb_size_cbf: 16
emb_size_edge: 512
emb_size_rbf: 16
emb_size_trip: 64
envelope:
exponent: 5
name: polynomial
extensive: true
max_neighbors: 50
num_after_skip: 2
num_atom: 3
num_before_skip: 1
num_blocks: 3
num_concat: 1
num_radial: 128
num_spherical: 7
otf_graph: false
output_init: HeOrthogonal
rbf:
name: gaussian
regress_forces: true
scale_file: configs/s2ef/all/gemnet/scaling_factors/gemnet-dT.json
noddp: false
optim:
batch_size: 1
clip_grad_norm: 10
ema_decay: 0.999
eval_batch_size: 1
factor: 0.8
force_coefficient: 100
lr_initial: 0.0005
max_epochs: 1
mode: min
num_workers: 2
optimizer: AdamW
optimizer_params:
amsgrad: true
patience: 3
scheduler: ReduceLROnPlateau
slurm: {}
task:
dataset: trajectory_lmdb
description: Regressing to energies and forces for DFT trajectories from OCP
eval_on_free_atoms: true
grad_input: atomic forces
labels:
- potential energy
metric: mae
num_relaxation_batches: 1
relax_dataset:
src: data/is2re/val_20/data.lmdb
relax_opt:
alpha: 70.0
damping: 1.0
maxstep: 0.04
memory: 50
traj_dir: ml-relaxations/is2rs-test
relaxation_steps: 200
train_on_free_atoms: true
type: regression
write_pos: true
trainer: forces
val_dataset:
src: data/s2ef/val_20
2022-10-31 18:01:20 (INFO): Batch balancing is disabled for single GPU training.
2022-10-31 18:01:20 (INFO): Batch balancing is disabled for single GPU training.
2022-10-31 18:01:20 (INFO): Loading dataset: trajectory_lmdb
2022-10-31 18:01:20 (INFO): Batch balancing is disabled for single GPU training.
2022-10-31 18:01:20 (INFO): Loading model: gemnet_t
/home/runner/micromamba-root/envs/buildenv/lib/python3.9/site-packages/torch/cuda/amp/grad_scaler.py:115: UserWarning: torch.cuda.amp.GradScaler is enabled, but CUDA is not available. Disabling.
warnings.warn("torch.cuda.amp.GradScaler is enabled, but CUDA is not available. Disabling.")
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In [6], line 1
----> 1 trainer = ForcesTrainer(
2 task=task,
3 model=model,
4 dataset=dataset,
5 optimizer=optimizer,
6 identifier="is2rs-example",
7 run_dir="./", # directory to save results if is_debug=False. Prediction files are saved here so be careful not to override!
8 is_debug=False, # if True, do not save checkpoint, logs, or results
9 print_every=5,
10 seed=0, # random seed to use
11 logger="tensorboard", # logger of choice (tensorboard and wandb supported)
12 local_rank=0,
13 amp=True, # use PyTorch Automatic Mixed Precision (faster training and less memory usage)
14 )
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/trainers/forces_trainer.py:88, in ForcesTrainer.__init__(self, task, model, dataset, optimizer, identifier, normalizer, timestamp_id, run_dir, is_debug, is_hpo, print_every, seed, logger, local_rank, amp, cpu, slurm, noddp)
67 def __init__(
68 self,
69 task,
(...)
86 noddp=False,
87 ):
---> 88 super().__init__(
89 task=task,
90 model=model,
91 dataset=dataset,
92 optimizer=optimizer,
93 identifier=identifier,
94 normalizer=normalizer,
95 timestamp_id=timestamp_id,
96 run_dir=run_dir,
97 is_debug=is_debug,
98 is_hpo=is_hpo,
99 print_every=print_every,
100 seed=seed,
101 logger=logger,
102 local_rank=local_rank,
103 amp=amp,
104 cpu=cpu,
105 name="s2ef",
106 slurm=slurm,
107 noddp=noddp,
108 )
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/trainers/base_trainer.py:205, in BaseTrainer.__init__(self, task, model, dataset, optimizer, identifier, normalizer, timestamp_id, run_dir, is_debug, is_hpo, print_every, seed, logger, local_rank, amp, cpu, name, slurm, noddp)
203 if distutils.is_master():
204 print(yaml.dump(self.config, default_flow_style=False))
--> 205 self.load()
207 self.evaluator = Evaluator(task=name)
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/trainers/base_trainer.py:214, in BaseTrainer.load(self)
212 self.load_datasets()
213 self.load_task()
--> 214 self.load_model()
215 self.load_loss()
216 self.load_optimizer()
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/trainers/base_trainer.py:369, in BaseTrainer.load_model(self)
364 bond_feat_dim = self.config["model_attributes"].get(
365 "num_gaussians", 50
366 )
368 loader = self.train_loader or self.val_loader or self.test_loader
--> 369 self.model = registry.get_model_class(self.config["model"])(
370 loader.dataset[0].x.shape[-1]
371 if loader
372 and hasattr(loader.dataset[0], "x")
373 and loader.dataset[0].x is not None
374 else None,
375 bond_feat_dim,
376 self.num_targets,
377 **self.config["model_attributes"],
378 ).to(self.device)
380 if distutils.is_master():
381 logging.info(
382 f"Loaded {self.model.__class__.__name__} with "
383 f"{self.model.num_params} parameters."
384 )
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/models/gemnet/gemnet.py:261, in GemNetT.__init__(self, num_atoms, bond_feat_dim, num_targets, num_spherical, num_radial, num_blocks, emb_size_atom, emb_size_edge, emb_size_trip, emb_size_rbf, emb_size_cbf, emb_size_bil_trip, num_before_skip, num_after_skip, num_concat, num_atom, regress_forces, direct_forces, cutoff, max_neighbors, rbf, envelope, cbf, extensive, otf_graph, use_pbc, output_init, activation, num_elements, scale_file)
252 self.int_blocks = torch.nn.ModuleList(int_blocks)
254 self.shared_parameters = [
255 (self.mlp_rbf3.linear.weight, self.num_blocks),
256 (self.mlp_cbf3.weight, self.num_blocks),
257 (self.mlp_rbf_h.linear.weight, self.num_blocks),
258 (self.mlp_rbf_out.linear.weight, self.num_blocks + 1),
259 ]
--> 261 load_scales_compat(self, scale_file)
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/modules/scaling/compat.py:55, in load_scales_compat(module, scale_file)
52 def load_scales_compat(
53 module: nn.Module, scale_file: Optional[Union[str, ScaleDict]]
54 ):
---> 55 scale_dict = _load_scale_dict(scale_file)
56 if not scale_dict:
57 return
File ~/work/ml_catalysis_tutorials/ml_catalysis_tutorials/ocp/ocpmodels/modules/scaling/compat.py:31, in _load_scale_dict(scale_file)
29 path = Path(scale_file)
30 if not path.exists():
---> 31 raise ValueError(f"Scale file {path} does not exist.")
33 scale_dict: Optional[ScaleDict] = None
34 if path.suffix == ".pt":
ValueError: Scale file configs/s2ef/all/gemnet/scaling_factors/gemnet-dT.json does not exist.
Load the best checkpoint#
trainer.load_checkpoint(checkpoint_path=checkpoint_path)
Run relaxations#
We run a full relaxation for a single batch of our relaxation dataset (num_relaxation_batches=1).
trainer.run_relaxations()
0%| | 0/20 [00:00<?, ?it/s]/content/ocp/ocpmodels/models/gemnet/gemnet.py:373: UserWarning: __floordiv__ is deprecated, and its behavior will change in a future version of pytorch. It currently rounds toward 0 (like the 'trunc' function NOT 'floor'). This results in incorrect rounding for negative values. To keep the current behavior, use torch.div(a, b, rounding_mode='trunc'), or for actual floor division, use torch.div(a, b, rounding_mode='floor').
neighbors_new // 2,
/content/ocp/ocpmodels/models/gemnet/gemnet.py:467: UserWarning: __floordiv__ is deprecated, and its behavior will change in a future version of pytorch. It currently rounds toward 0 (like the 'trunc' function NOT 'floor'). This results in incorrect rounding for negative values. To keep the current behavior, use torch.div(a, b, rounding_mode='trunc'), or for actual floor division, use torch.div(a, b, rounding_mode='floor').
block_sizes = neighbors // 2
5%|▌ | 1/20 [00:10<03:23, 10.72s/it]
Visualize ML-driven relaxations#
Following our earlier visualization steps, we can plot our ML-generated relaxations.
import glob
import random
import ase.io
import matplotlib
import matplotlib.pyplot as plt
from ase.visualize.plot import plot_atoms
params = {
"axes.labelsize": 14,
"font.size": 14,
"font.family": " DejaVu Sans",
"legend.fontsize": 20,
"xtick.labelsize": 20,
"ytick.labelsize": 20,
"axes.labelsize": 25,
"axes.titlesize": 25,
"text.usetex": False,
"figure.figsize": [12, 12],
}
matplotlib.rcParams.update(params)
system = glob.glob("ml-relaxations/is2rs-test/*.traj")[0]
ml_trajectory = ase.io.read(system, ":")
energies = [atom.get_potential_energy() for atom in ml_trajectory]
plt.figure(figsize=(7, 5))
plt.plot(range(len(energies)), energies)
plt.xlabel("step")
plt.ylabel("energy, eV")
system
'ml-relaxations/is2rs-test/1700380.traj'
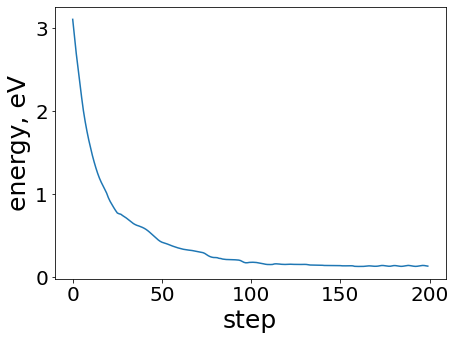
Qualitatively, the ML relaxation is behaving as expected - decreasing energies over the course of the relaxation.
fig, ax = plt.subplots(1, 3)
labels = ["ml-initial", "ml-middle", "ml-final"]
for i in range(3):
ax[i].axis("off")
ax[i].set_title(labels[i])
ase.visualize.plot.plot_atoms(
ml_trajectory[0],
ax[0],
radii=0.8,
# rotation=("-75x, 45y, 10z")) # uncomment to visualize at different angles
)
ase.visualize.plot.plot_atoms(
ml_trajectory[100],
ax[1],
radii=0.8,
# rotation=("-75x, 45y, 10z") # uncomment to visualize at different angles
)
ase.visualize.plot.plot_atoms(
ml_trajectory[-1],
ax[2],
radii=0.8,
# rotation=("-75x, 45y, 10z"), # uncomment to visualize at different angles
)
<AxesSubplot: title={'center': 'ml-final'}>
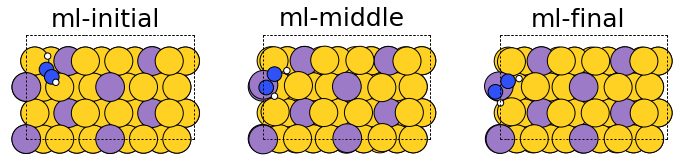
Qualitatively, the generated structures seem reasonable with no obvious issues we had previously mentioned to look out for.

