HW1 (due 9/4)
Contents
HW1 (due 9/4)#
Catalyst deactivation in a batch reactor [50 pts]#
Consider the irreversible, liquid-phase isomerization reaction carried out in a solvent containing dissolved catalyst at 25 C in a batch reactor:
The apparent first-order reaction rate constant \(k_b\) decreases with time because of catalyst deterioriation. A chemist friend of yours has studied the catalyst deactivation process and has proposed that it can be modeled with
in which \(k\) is the fresh catalyst rate constant and \(k_d\) is the deactivation rate constant.
We can derive the ODE to solve by starting with a mol balance on the entire system:
with the initial condition \(C_b(t=0)=C_{b0}=5\) M.
Mol balance solve#
Solve the mole balance for \(C_B(t)\) assuming \(k\)=0.6/hr and \(k_d\)=2/hr for the first two hours. Plot the conversion % for your solution (defined as \(1-C_B(t)/C_{B0}\)).
import matplotlib.pyplot as plt
import numpy as np
from scipy.integrate import solve_ivp
# define parameters
k = 0.6 # 1/hr
k_d = 2.0 # 1/hr
Cb_0 = 5 # M
# define the ODE
def dCb_dt(t: float, Cb: float) -> float:
return -(k * Cb) / (1 + k_d * t)
# define time span and eval times
t_span = [0, 5]
t_eval = np.linspace(*t_span, 50)
# integrate ODE
sol = solve_ivp(fun=dCb_dt, t_span=t_span, y0=[Cb_0], t_eval=t_eval)
# get Cb(t)
Cb_t = sol.y[0]
# compute conversion %
conv = 1 - Cb_t / Cb_0
# plot conversion
fig, ax = plt.subplots()
ax.plot(t_eval, conv)
ax.set_xlabel("t [hr]")
ax.set_ylabel(r"conversion %")
Text(0, 0.5, 'conversion %')
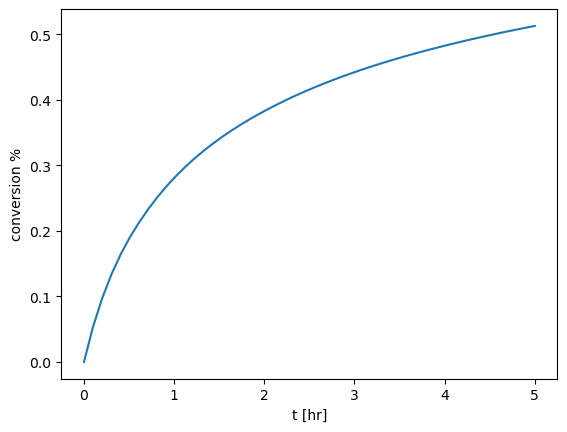
If it takes two hours to reach 50% conversion and the fresh catalyst has a rate constant of 0.6/hr what is the actual \(k_d\)?#
We know that \(C_b(t=0)=C_{b0}\), \(\frac{C_b(t=2)}{C_{b0}} = 0.5\) and \(k = 0.6/hr\). We attempt to solve the ODE analytically to find an expression for \(C_b(t)\):
To solve for \(k_d\), we use scipy.optimize.fsolve:
from scipy.optimize import fsolve
# define parameters
t = 2.0 # h
conv_at_t = 0.5 # C_b(t=2)/C_b0
k = 0.6 # 1/hr
# ODE function to integrate
def dCb_dt(t: float, Cb: float, k_d: float) -> float:
return -(k * Cb) / (1 + k_d * t)
# Objective to decide how close the solution is for a given k_d
# We want to make this objective equal to 0!
def objective(k_d):
# Solve the ODE
sol = solve_ivp(fun=dCb_dt, t_span=[0, 2], y0=[Cb_0], args=(k_d,))
# Get the final concentration
Cb = sol.y[0, -1]
# Return how far the final conversion is from 0.5
return (1 - (Cb / Cb_0)) - 0.5
k_d_actual = fsolve(func=objective, x0=[2])[0]
print(f"The actual k_d is {k_d_actual:.3f}")
The actual k_d is 0.877
Using \(k_d\) you found from the previous step, use solve_ivp events to determine how long it takes to reach 75% conversion in the reactor.#
# create new event for conversion = 0.75
def conversionTargetReached(t: float, Cb: np.array, k_d: float) -> float:
return (1 - (Cb[0] / Cb_0)) - 0.75
conversionTargetReached.terminal = True
# set longer time span
t_span = [0, 10]
t_eval = np.linspace(*t_span, 50)
# integrate ODE with event
sol = solve_ivp(
fun=dCb_dt,
t_span=t_span,
y0=[Cb_0],
t_eval=t_eval,
args=(k_d_actual,),
events=conversionTargetReached,
)
# print time of event
print("Conversion target hit at: " + str(sol.t_events[0][0]) + " h.")
Conversion target hit at: 7.508500707147827 h.
Catalyst refresh#
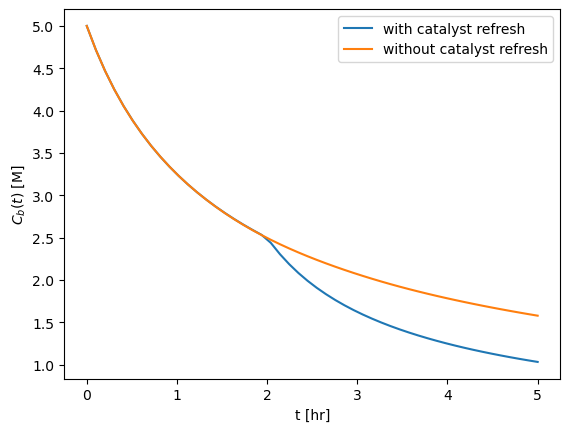
Say that we can stop the batch process after 2 hours, filter the spent catalyst, and replace with fresh catalyst. \(C_B\) will start wherever the first reaction left off. Solve and plot for \(C_B(t)\) over 5 hours, and include the plot with the catalyst refresh. Include a legend on the plot to label each solution
# we define a piecewise function to model the catalyst refresh
def dCb_dt_refresh(t: float, Cb: float, refresh: bool = True) -> float:
if t > 2.0 and refresh is True:
k_b = k / (1 + k_d_actual * (t - 2.0))
else:
k_b = k / (1 + k_d_actual * t)
return -k_b * Cb
# set longer time span
t_span = [0, 5]
t_eval = np.linspace(*t_span, 50)
# integrate with refresh
# Note: k_b(t) is non-smooth -> use implicit method
sol_with_refresh = solve_ivp(
fun=dCb_dt_refresh,
t_span=t_span,
y0=[Cb_0],
t_eval=t_eval,
args=(True,),
method="Radau",
)
Cb_with_refresh = sol_with_refresh.y[0]
# integrate without refresh
sol_without_refresh = solve_ivp(
fun=dCb_dt_refresh, t_span=t_span, y0=[Cb_0], t_eval=t_eval, args=(False,)
)
Cb_without_refresh = sol_without_refresh.y[0]
# plot concentrations over time
fig, ax = plt.subplots()
ax.plot(t_eval, Cb_with_refresh, label="with catalyst refresh")
ax.plot(t_eval, Cb_without_refresh, label="without catalyst refresh")
ax.set_xlabel("t [hr]")
ax.set_ylabel(r"$C_b(t)$ [M]")
ax.legend()
<matplotlib.legend.Legend at 0x7f35d7a6aac0>
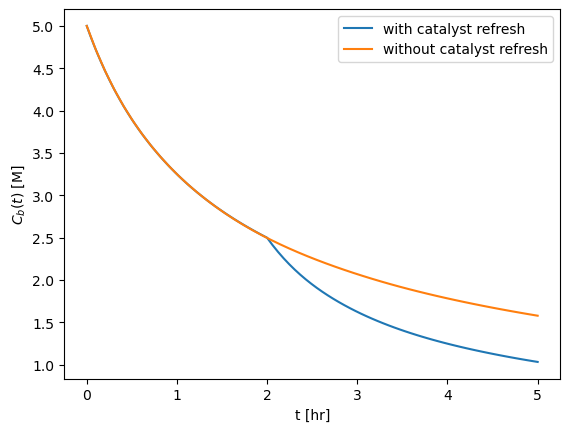
We could also do this with two different ODE solves! Both give the same answer.
# we define a piecewise function to model the catalyst refresh
def dCb_dt(t: float, Cb: float) -> float:
k_b = k / (1 + k_d_actual * t)
return -k_b * Cb
# integrate before refresh
sol_before_refresh = solve_ivp(
fun=dCb_dt,
t_span=[0, 2],
y0=[Cb_0],
t_eval=np.linspace(0, 2),
)
# Integrate after refresh using the final concentration from the previous step
sol_after_refresh = solve_ivp(
fun=dCb_dt,
t_span=[0, 3],
y0=[sol_before_refresh.y[0, -1]],
t_eval=np.linspace(0, 3),
)
# Combine the times and concentrations into one solution
# Note that the second time has 2 added to it!
Cb_with_refresh = np.hstack((sol_before_refresh.y, sol_after_refresh.y)).T
t_eval_with_refresh = np.hstack((sol_before_refresh.t, 2 + sol_after_refresh.t))
# Integrate without refresh
sol_without_refresh = solve_ivp(fun=dCb_dt, t_span=t_span, y0=[Cb_0], t_eval=t_eval)
Cb_without_refresh = sol_without_refresh.y[0]
# plot concentrations over time
fig, ax = plt.subplots()
ax.plot(t_eval_with_refresh, Cb_with_refresh, label="with catalyst refresh")
ax.plot(t_eval, Cb_without_refresh, label="without catalyst refresh")
ax.set_xlabel("t [hr]")
ax.set_ylabel(r"$C_b(t)$ [M]")
ax.legend()
<matplotlib.legend.Legend at 0x7f35d79356d0>
Curve fitting with two species and multiple data (50 pt)#
Consider the irreversible reaction:
with \(r=kC_A^nC_B^m\) taking place in an isothermal liquid-phase batch reactor. Measurements of \(C_A\) vs \(C_B\) are included in the attached file isothermal_liquidphase_batch_data.dat. We wish to determine from the data the rate constant and the order of the reaction with respect to A and B. We have data from two experiments.
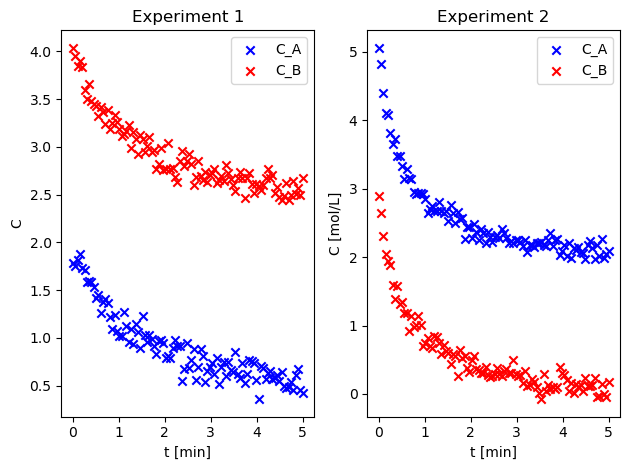
Load the data from the file into a numpy array and plot the concentration of each species#
You can use either the csv library https://docs.python.org/3/library/csv.html or pandas https://pandas.pydata.org/pandas-docs/stable/generated/pandas.read_csv.html.
The first column is time in minutes. The second and third column is C_A and C_B for the first experiment (in mol/L). The fourth and fifth column is C_A and C_B for the second experiment. Plot the data for \(C_A\) and \(C_B\) for each experiment (one experiment per figure).
import matplotlib.pyplot as plt
import pandas as pd
# read data using regular expression separator
df = pd.read_csv(
"isothermal_liquidphase_batch.dat",
sep=r"\s{2,}",
names=["t", "C_A1", "C_B1", "C_A2", "C_B2"],
engine="python",
)
# divide data into two data frames with same columns
cols = ["t", "C_A", "C_B"]
df_exp1 = df[["t", "C_A1", "C_B1"]]
df_exp1.columns = cols
df_exp2 = df[["t", "C_A2", "C_B2"]]
df_exp2.columns = cols
# plot data from experiment 1
fig, ax = plt.subplots(1, 2)
# df_exp1.plot(x="t", ax=ax1)
ax[0].scatter(
x=df_exp1[["t"]].values,
y=df_exp1[["C_A"]].values,
marker="x",
color="b",
label="C_A",
)
ax[0].scatter(
x=df_exp1[["t"]].values,
y=df_exp1[["C_B"]].values,
marker="x",
color="r",
label="C_B",
)
ax[0].set_ylabel("C")
ax[0].set_xlabel("t [min]")
ax[0].set_title("Experiment 1")
ax[0].legend()
# plot data from experiment 2
ax[1].scatter(
x=df_exp2[["t"]].values,
y=df_exp2[["C_A"]].values,
marker="x",
color="b",
label="C_A",
)
ax[1].scatter(
x=df_exp2[["t"]].values,
y=df_exp2[["C_B"]].values,
marker="x",
color="r",
label="C_B",
)
ax[1].set_ylabel("C [mol/L]")
ax[1].set_xlabel("t [min]")
ax[1].set_title("Experiment 2")
ax[1].legend()
fig.tight_layout()
Using lmfit, estimate rate parameters \(k, n, m\) and initial concentration \(C_{A0},C_{B0}\) from the data in the first experiment using a numerical solution for the concentrations of each species (standard mol balance + odeint). Plot the final fit of the experiment along with the experimental data and calculate the uncertainty in each value#
import numpy as np
from lmfit import Model
from scipy.integrate import odeint, solve_ivp
# define ODe for ivp
def reaction_ODE_ivp(t, C, k, n, m):
C_A, C_B = C
r = k * (C_A**n) * (C_B**m)
dC_A_dt = -r
dC_B_dt = -r
return np.array([dC_A_dt, dC_B_dt])
# integrate ODE using ivp
def integrate_ODE(t, C_A0, C_B0, k, n, m, method):
sol = solve_ivp(
reaction_ODE_ivp,
t_span=(0, t[-1]),
t_eval=t,
y0=np.array([C_A0, C_B0]),
args=(k, n, m),
method=method,
)
C = sol.y.T
return C
# define model and fit to data
def fit_model_to_data(t, C_A, C_B, integration_method="RK45"):
data = np.concatenate([C_A, C_B], axis=1)
gmodel = Model(
integrate_ODE,
independent_vars=["t"],
param_names=["C_A0", "C_B0", "k", "n", "m"],
method=integration_method,
)
params = gmodel.make_params(C_A0=C_A[0, 0], C_B0=C_B[0, 0], k=2.0, n=2.0, m=2.0)
result = gmodel.fit(data=data, params=params, t=t)
return result
# plot data and predictions
def plot_result(result, t, C_A, C_B, ax, title):
prediction = result.eval(t=t)
# plot data from experiment
ax.scatter(t, C_A, label="Data $C_A$", marker="x", color="b")
ax.scatter(t, C_B, label="Data $C_B$", marker="x", color="r")
# plot predictions
ax.plot(t, prediction[:, 0], label="Prediction", color="k")
ax.plot(t, prediction[:, 1], color="k")
ax.set_ylabel("C [mol/L]")
ax.set_xlabel("t [min]")
ax.set_title(title)
ax.legend()
# get data as numpy arrays
t = df["t"].values
C_A1 = df["C_A1"].values.reshape(-1, 1)
C_B1 = df["C_B1"].values.reshape(-1, 1)
result1 = fit_model_to_data(t, C_A1, C_B1)
t_eval = t
# plot data from experiment 1
fig, ax = plt.subplots()
plot_result(result1, t_eval, C_A1, C_B1, ax, "Experiment 1")
# print model info
print(result1.fit_report())
[[Model]]
Model(integrate_ODE, method='RK45')
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 613
# data points = 200
# variables = 5
chi-square = 1.74113683
reduced chi-square = 0.00892891
Akaike info crit = -938.755824
Bayesian info crit = -922.264237
[[Variables]]
C_A0: 1.95374598 +/- 0.04451124 (2.28%) (init = 1.785)
C_B0: 3.96043184 +/- 0.04452466 (1.12%) (init = 4.036)
k: 3.26348562 +/- 17.0540000 (522.57%) (init = 2)
n: 3.15625683 +/- 1.64028660 (51.97%) (init = 2)
m: -2.16720723 +/- 4.74031974 (218.73%) (init = 2)
[[Correlations]] (unreported correlations are < 0.100)
C(k, m) = -1.000
C(n, m) = -0.997
C(k, n) = 0.997
C(C_A0, C_B0) = 0.955
C(C_B0, m) = 0.584
C(C_B0, k) = -0.583
C(C_A0, m) = 0.579
C(C_A0, k) = -0.579
C(C_B0, n) = -0.550
C(C_A0, n) = -0.540
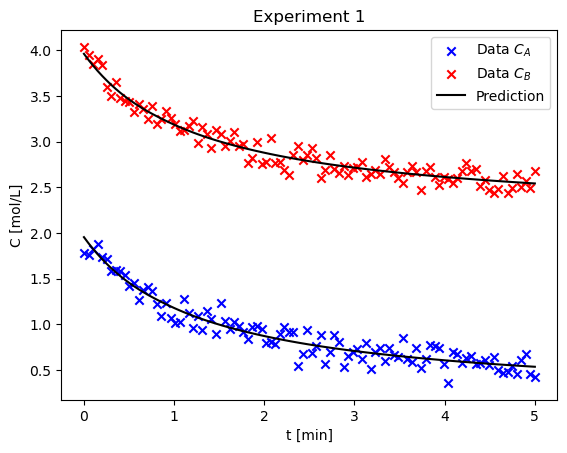
Consider the second experiment, estimate the parameters using only this data. Comment on how the values and confidence intervals are different than the first case#
# get data as numpy arrays
t = df["t"].values
C_A2 = df["C_A2"].values.reshape(-1, 1)
C_B2 = df["C_B2"].values.reshape(-1, 1)
result2 = fit_model_to_data(t, C_A2, C_B2, integration_method="RK45")
# plot data from experiment 2
fig, ax = plt.subplots()
plot_result(result2, t_eval, C_A2, C_B2, ax, "Experiment 2")
# print model info
print(result2.fit_report())
[[Model]]
Model(integrate_ODE, method='RK45')
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 194
# data points = 200
# variables = 5
chi-square = 1.68848917
reduced chi-square = 0.00865892
Akaike info crit = -944.896644
Bayesian info crit = -928.405057
[[Variables]]
C_A0: 5.02591001 +/- 0.05648774 (1.12%) (init = 5.059)
C_B0: 3.01505138 +/- 0.05636400 (1.87%) (init = 2.888)
k: 0.03315831 +/- 0.01859468 (56.08%) (init = 2)
n: 2.98279103 +/- 0.48681468 (16.32%) (init = 2)
m: 0.69365669 +/- 0.14775385 (21.30%) (init = 2)
[[Correlations]] (unreported correlations are < 0.100)
C(k, n) = -1.000
C(n, m) = -0.979
C(k, m) = 0.979
C(C_A0, C_B0) = 0.973
C(C_A0, n) = 0.440
C(C_A0, k) = -0.430
C(C_B0, n) = 0.419
C(C_B0, k) = -0.409
C(C_A0, m) = -0.354
C(C_B0, m) = -0.324
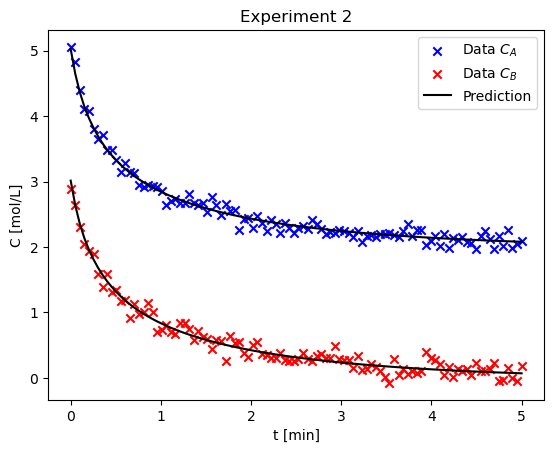
Estimate the parameters using both experiments simultaneously. Are the confidence intervals better? Comment on why or why not.#
# integrate both ODEs using respective initial conditions
def integrate_ODE_both(t, C_A0_1, C_B0_1, C_A0_2, C_B0_2, k, n, m, method):
C_1 = solve_ivp(
reaction_ODE_ivp,
t_span=(0, t[-1]),
t_eval=t,
y0=np.array([C_A0_1, C_B0_1]),
args=(k, n, m),
method=method[0],
).y.T
C_2 = solve_ivp(
reaction_ODE_ivp,
t_span=(0, t[-1]),
t_eval=t,
y0=np.array([C_A0_2, C_B0_2]),
args=(k, n, m),
method=method[1],
).y.T
return np.concatenate([C_1, C_2], axis=1)
# define and fit model
def fit_model_to_data_both(
t, C_A_1, C_B_1, C_A_2, C_B_2, integration_method=["RK45", "RK45"]
):
# define data set
data = np.concatenate([C_A_1, C_B_1, C_A_2, C_B_2], axis=1)
# define model
gmodel = Model(
integrate_ODE_both,
independent_vars=["t"],
param_names=["C_A0_1", "C_B0_1", "C_A0_2", "C_B0_2", "k", "n", "m"],
method=integration_method,
)
params = gmodel.make_params(
# initial guesses - very sensitive
C_A0_1=2,
C_B0_1=4,
C_A0_2=5,
C_B0_2=3,
k=2,
n=2,
m=2,
method=integration_method,
)
result = gmodel.fit(data=data, params=params, t=t)
return result
# plot predictions and data
def plot_results_both(results_both, t, C_A1, C_B1, C_A2, C_B2, ax, title):
prediction = result_both.eval(t=t)
ax[0].scatter(t, C_A1, label="Data $C_A$", marker="x", color="b")
ax[0].scatter(t, C_B1, label="Data $C_B$", marker="x", color="r")
ax[0].plot(t, prediction[:, 0], label="Prediction", color="k")
ax[0].plot(t, prediction[:, 1], color="k")
ax[0].legend()
ax[0].set_ylabel("C [mol/L]")
ax[0].set_xlabel("t [min]")
ax[0].set_title("Experiment 1 " + title)
ax[1].scatter(t, C_A2, label="Data $C_A$", marker="x", color="b")
ax[1].scatter(t, C_B2, label="Data $C_B$", marker="x", color="r")
ax[1].plot(t, prediction[:, 2], label="Prediction", color="k")
ax[1].plot(t, prediction[:, 3], color="k")
ax[1].legend()
ax[1].set_ylabel("C [mol/L]")
ax[1].set_xlabel("t [min]")
ax[1].set_title("Experiment 2 " + title)
# fit model
result_both = fit_model_to_data_both(
t, C_A1, C_B1, C_A2, C_B2, integration_method=["RK45", "RK45"]
)
# plot data from both experiments
fig, ax = plt.subplots(1, 2)
plot_results_both(result_both, t, C_A1, C_B1, C_A2, C_B2, ax, "noisy")
fig.tight_layout()
# print model info
print(result_both.fit_report())
[[Model]]
Model(integrate_ODE_both, method='['RK45', 'RK45']')
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 175
# data points = 400
# variables = 7
chi-square = 3.46646955
reduced chi-square = 0.00882053
Akaike info crit = -1885.33116
Bayesian info crit = -1857.39091
[[Variables]]
C_A0_1: 1.96326640 +/- 0.02921224 (1.49%) (init = 2)
C_B0_1: 3.96989620 +/- 0.02957283 (0.74%) (init = 4)
C_A0_2: 4.98419433 +/- 0.04920060 (0.99%) (init = 5)
C_B0_2: 2.97642345 +/- 0.04963032 (1.67%) (init = 3)
k: 0.09838761 +/- 0.00342137 (3.48%) (init = 2)
n: 2.03729617 +/- 0.03103060 (1.52%) (init = 2)
m: 0.98457641 +/- 0.02522301 (2.56%) (init = 2)
[[Correlations]] (unreported correlations are < 0.100)
C(C_A0_2, C_B0_2) = 0.965
C(C_A0_1, C_B0_1) = 0.898
C(k, n) = -0.893
C(k, m) = -0.697
C(n, m) = 0.572
C(C_B0_2, n) = 0.524
C(C_A0_2, n) = 0.500
C(C_B0_2, m) = 0.413
C(C_B0_2, k) = -0.370
C(C_A0_2, m) = 0.343
C(C_B0_1, m) = 0.333
C(C_A0_2, k) = -0.312
C(C_A0_1, m) = 0.300
C(C_A0_1, C_B0_2) = 0.146
C(C_B0_1, C_B0_2) = 0.137
C(C_A0_1, C_A0_2) = 0.135
C(C_B0_1, C_A0_2) = 0.127
C(C_B0_1, k) = 0.116
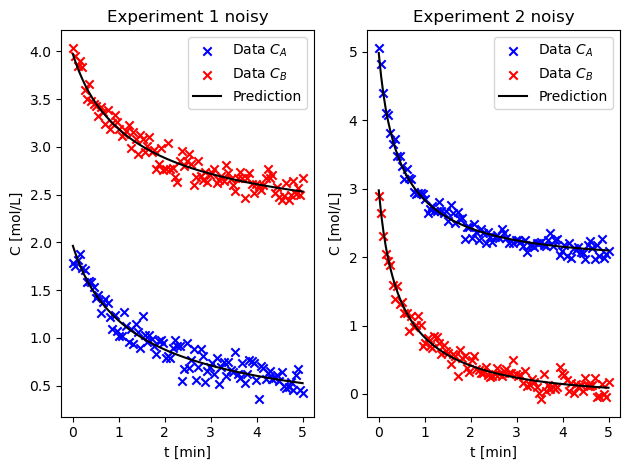
Using the noisy data in isothermal_liquidphase_batch_errorfree.dat, repeat the fit using all of the data. Do the results agree with your initial trials?#
# load noise-free data
df_ef = pd.read_csv(
"isothermal_liquidphase_batch_errorfree.dat",
sep=r"\s{2,}",
names=["t", "C_A1", "C_B1", "C_A2", "C_B2"],
engine="python",
)
# divide data into two data frames with same columns
cols = ["t", "C_A", "C_B"]
df_exp1_ef = df_ef[["t", "C_A1", "C_B1"]]
df_exp1_ef.columns = cols
df_exp2_ef = df_ef[["t", "C_A2", "C_B2"]]
df_exp2_ef.columns = cols
t_ef = df_ef["t"].values
C_A1_ef = df_ef["C_A1"].values.reshape(-1, 1)
C_B1_ef = df_ef["C_B1"].values.reshape(-1, 1)
C_A2_ef = df_ef["C_A2"].values.reshape(-1, 1)
C_B2_ef = df_ef["C_B2"].values.reshape(-1, 1)
# fit model to data
result_both_ef = fit_model_to_data_both(
t_ef, C_A1_ef, C_B1_ef, C_A2_ef, C_B2_ef, integration_method=["RK45", "RK45"]
)
# plot data from both experiments
fig_ef, ax_ef = plt.subplots(1, 2)
plot_results_both(
result_both_ef, t_ef, C_A1_ef, C_B1_ef, C_A2_ef, C_B2_ef, ax_ef, "noise-free"
)
fig.tight_layout()
# print model info
print(result_both_ef.fit_report())
[[Model]]
Model(integrate_ODE_both, method='['RK45', 'RK45']')
[[Fit Statistics]]
# fitting method = leastsq
# function evals = 232
# data points = 400
# variables = 7
chi-square = 3.0180e-04
reduced chi-square = 7.6793e-07
Akaike info crit = -5624.88794
Bayesian info crit = -5596.94769
[[Variables]]
C_A0_1: 2.00087760 +/- 2.7655e-04 (0.01%) (init = 2)
C_B0_1: 4.00093588 +/- 2.7998e-04 (0.01%) (init = 4)
C_A0_2: 5.00230800 +/- 4.5646e-04 (0.01%) (init = 5)
C_B0_2: 3.00237988 +/- 4.6023e-04 (0.02%) (init = 3)
k: 0.10032598 +/- 3.2176e-05 (0.03%) (init = 2)
n: 1.99989908 +/- 2.8352e-04 (0.01%) (init = 2)
m: 1.00227331 +/- 2.3422e-04 (0.02%) (init = 2)
[[Correlations]] (unreported correlations are < 0.100)
C(C_A0_2, C_B0_2) = 0.964
C(C_A0_1, C_B0_1) = 0.901
C(k, n) = -0.890
C(k, m) = -0.704
C(n, m) = 0.571
C(C_B0_2, n) = 0.523
C(C_A0_2, n) = 0.501
C(C_B0_2, m) = 0.412
C(C_B0_2, k) = -0.371
C(C_A0_2, m) = 0.341
C(C_B0_1, m) = 0.339
C(C_A0_2, k) = -0.312
C(C_A0_1, m) = 0.307
C(C_A0_1, C_B0_2) = 0.152
C(C_B0_1, C_B0_2) = 0.143
C(C_A0_1, C_A0_2) = 0.141
C(C_B0_1, C_A0_2) = 0.132
C(C_B0_1, k) = 0.100
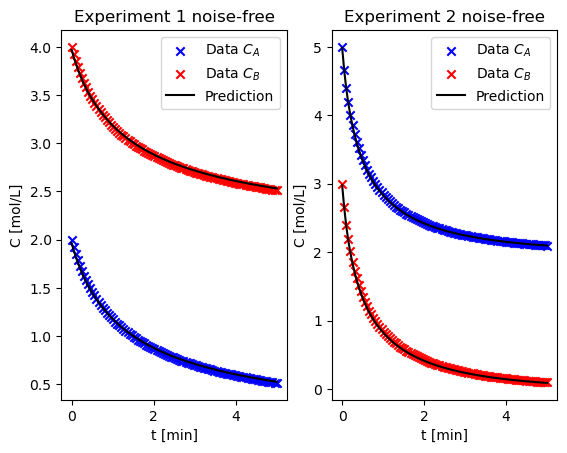
Based on these results, to determine the order of the rate expression with respect to two different species, how should you choose the initial conditions of the two species?#
The parameters should be chosen according to the optimal solutions computed using the full data set.



